dbGSS IDs and GenBank Accession Nos.of STSs
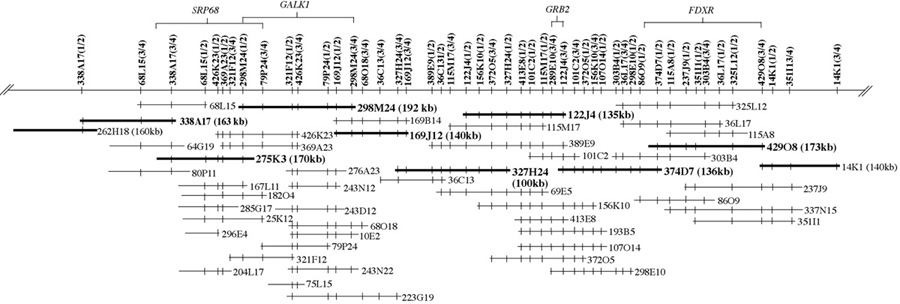
Figure 2. (more details) Schematic representation of a physical map of 50 BAC clones and 49 new STSs generated and assembled into a contig with an average depth of 6.4 BACs per STS marker. The STS name reveals which primer pair was used to create the STS. For example STS 338A17(1/2) was created utilizing primers 338A17-1 and 338A17-2 designed from direct end sequencing of the BAC 338A17. The primer sequences, PCR conditions and GenBank accession numbers are detailed by STS marker name (see links above). Clones that form a minimal tiling path across ~1.5 Mb are shown with a thicker line (not drawn to scale). Low-pass 3.2x sequence of ~1.2 Mb from eight BAC clones (in bold) from the minimal tiling path has been generated and analyzed. Brackets above the contigs identify BACs were canine/human sequence homologies were identified for genes SRP68, GALK1, GRB2, FDXR. The GRB2 gene was identified in two BACs (374D7 and 122J4) suggesting that canine GRB2 is located in overlapping regions between these. The gene order for SRP68, GALK1, GRB2, FDXR on the physical map confirms the gene order initially established on the CFA9-RH083000 map.