These 3 slides were copied from a larger slide set at http://www3.kumc.edu/jcalvet/bioc801b/index.htm
, They are owned by Pr. James P. Calvet at the Department of Biochemistry
and Molecular Biology in the University of Kansas Medical Center.
They are a nice set of slides that you could visit as well, but for our
purposes, these three slides suffice.
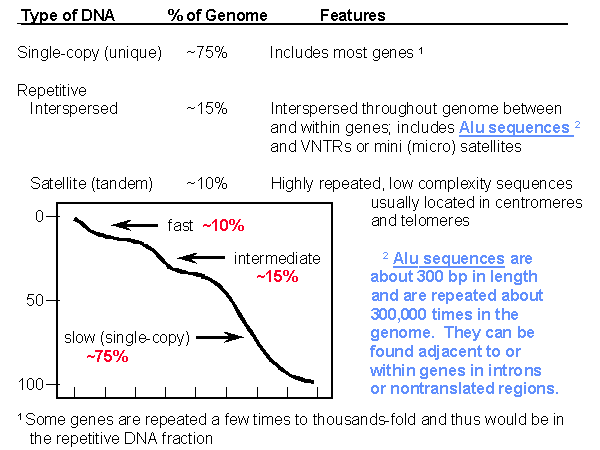
The human genome consists
of three populations of DNA: the fast and intermediate fractions make up
about 10% and 15% of the genome, respectively, and the slow fraction makes
up about 75% of the genome. Most of the genes in the human genome
are in the single-copy fraction. Repeated sequences can be of two
types: those that are interspersed throughout the genome or those that
are tandemly repeated satellite DNAs. Among the interspersed
repetitive sequences are so-called "Alu" sequences, which are about 300
base pairs in length and are repeated about 300,000 times in the genome.
They can be found adjacent to or within genes, and as illustrated later,
their presence can sometimes lead to the occasional disruption of genes.
The interspersed repetitive sequences also include VNTRs (variable numbers
of tandem repeats), which are comprised of short repeated sequences of
only a few base-pairs, but of variable lengths. They, too, are interspersed
throughout the genome, and are quite useful as landmarks for mapping genes.
please close this window