Optimization of the UNRES Force Field by Hierarchical Design of the Potential-Energy Landscape. 2. Off-Lattice Tests of the Method with Single Proteins
 aw O
aw O dziej,
dziej,



 aw Pillardy,
aw Pillardy,
 and
and 
Baker Laboratory of Chemistry and Chemical Biology and Cornell Theory
Center, Cornell University, Ithaca, New York 14853-1301, and Faculty of
Chemistry, University of Gda sk, Sobieskiego 18, 80-952 Gda
sk, Sobieskiego 18, 80-952 Gda sk,
Poland
sk,
Poland
Received: April 30, 2004
In Final Form: August 12, 2004
Abstract:
We describe the application of our recently proposed method of hierarchical
optimization of the protein energy landscape to optimize our off-lattice
united-residue (UNRES) force field using single training proteins. First, the
IgG-binding domain from streptococcal protein G (PDB code 1IGD) was treated;
earlier attempts to use this protein to optimize the force field by optimizing
the energy gap and Z score between the nativelike and non-native
structures failed. The structure of this protein consists of an N-terminal
antiparallel 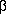 -hairpin, a middle
-hairpin, a middle
 -helix, and a C-terminal antiparallel
-helix, and a C-terminal antiparallel
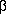 -hairpin, these elements being
referred to as
-hairpin, these elements being
referred to as 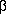 1,
1,
 2, and
2, and
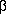 3, respectively, with
the two hairpins forming a parallel
3, respectively, with
the two hairpins forming a parallel
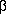 -sheet packed against the
-sheet packed against the
 -helix. In our earlier study, one of these
elements was assumed to form at level 1, two at level 2, and three at level 3,
and higher levels corresponded to the proper packing of two or more elements.
This approach resulted in a structure with the wrong packing of the
-helix. In our earlier study, one of these
elements was assumed to form at level 1, two at level 2, and three at level 3,
and higher levels corresponded to the proper packing of two or more elements.
This approach resulted in a structure with the wrong packing of the
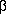 -sheet, and attempts at further
optimization failed. We therefore tried a hierarchy scheme that corresponds to
the sequence of folding events deduced from NMR experiments. In this scheme,
level 1 corresponds to structures with either
-sheet, and attempts at further
optimization failed. We therefore tried a hierarchy scheme that corresponds to
the sequence of folding events deduced from NMR experiments. In this scheme,
level 1 corresponds to structures with either
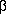 3 or
3 or
 2, level 2 to structures with both
2, level 2 to structures with both
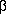 3 and
3 and
 2, level 3 to structures with
2, level 3 to structures with
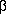 3,
3,
 2, and the N-terminal strand packed
against
2, and the N-terminal strand packed
against  2 (with
2 (with
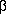 1 still not fully
formed), and level 4 to structures with
1 still not fully
formed), and level 4 to structures with
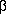 1,
1,
 2, and
2, and
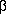 3, with
3, with
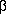 3 being packed to
3 being packed to
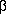 1, which also implies
the packing of
1, which also implies
the packing of 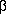 1 and
1 and
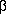 3 against
3 against
 2. This optimization was successful
and resulted in a reasonably transferable force field that led to well-foldable
proteins. This corroborates the conclusion from our model on-lattice studies (Liwo,
A.; Ar
2. This optimization was successful
and resulted in a reasonably transferable force field that led to well-foldable
proteins. This corroborates the conclusion from our model on-lattice studies (Liwo,
A.; Ar ukowicz, P.; O
ukowicz, P.; O dziej, S.;
Czaplewski, C.; Makowski, M.; Scheraga, H. A. J. Phys. Chem. B 2004,
108, 16918) that a proper design of the structural hierarchy is of
crucial importance to the foldability with the resulting potential-energy
function. Moreover, in the off-lattice approach, the design of the hierarchy
also appears to be important to the success of the optimization procedure
itself. The next series of calculations was carried out with the LysM domain
from the E. coli 1E0G (
dziej, S.;
Czaplewski, C.; Makowski, M.; Scheraga, H. A. J. Phys. Chem. B 2004,
108, 16918) that a proper design of the structural hierarchy is of
crucial importance to the foldability with the resulting potential-energy
function. Moreover, in the off-lattice approach, the design of the hierarchy
also appears to be important to the success of the optimization procedure
itself. The next series of calculations was carried out with the LysM domain
from the E. coli 1E0G ( +
+
 ) protein, which is smaller than
1IGD. In this case, no experimental information about the folding pathway is
available; nevertheless, we were able to deduce the appropriate hierarchy by a
trial-and-error method. The resulting force field performed worse in tests on
) protein, which is smaller than
1IGD. In this case, no experimental information about the folding pathway is
available; nevertheless, we were able to deduce the appropriate hierarchy by a
trial-and-error method. The resulting force field performed worse in tests on
 +
+
 - and
- and
 -proteins than that derived on
the basis of 1IGD with a correct hierarchy, which suggests that the structure of
the 1IGD protein encodes more structure-determining interactions common to all
proteins than the 1E0G protein does. For 1E0G, we also attempted to carry out a
single energy gap and Z-score optimization; this effort resulted in an
unsearchable force field. (The nativelike structures could not be found by a
global search, although they were the lowest in energy). Technical details of
the method, including the maintenance of proper secondary structure and a method
to classify structure, are also described.
-proteins than that derived on
the basis of 1IGD with a correct hierarchy, which suggests that the structure of
the 1IGD protein encodes more structure-determining interactions common to all
proteins than the 1E0G protein does. For 1E0G, we also attempted to carry out a
single energy gap and Z-score optimization; this effort resulted in an
unsearchable force field. (The nativelike structures could not be found by a
global search, although they were the lowest in energy). Technical details of
the method, including the maintenance of proper secondary structure and a method
to classify structure, are also described.